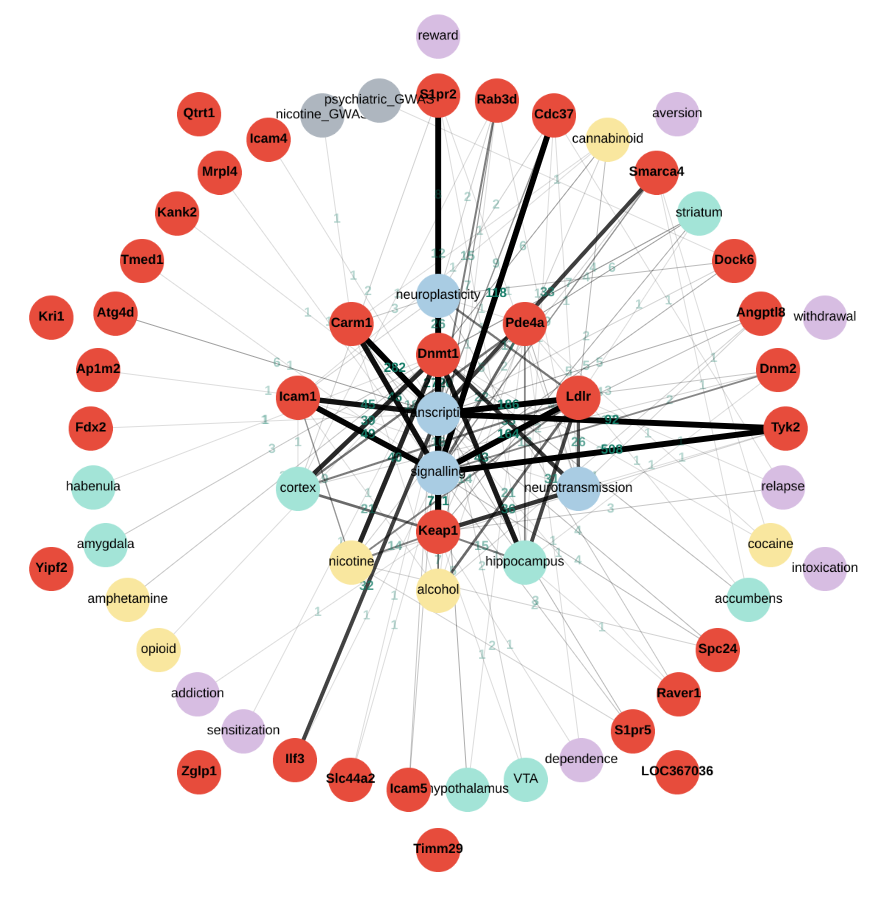
Mining PubMed for Gene Relationships using Custom Ontologies
GeneCup is a tool to answer the question "What do we know about these genes and the topic I study?". GeneCup answers this question by searching PubMed to find sentences containing gene symbols and a custom list of keywords. Human GWAS findings from NHGRI-EBI GWAS catalog are also included in the search. These gene-keyword relationships are presented as a graph or a table. Here we present a curated ontology for drug addiction with over 300 keywords organized into seven categories. Deep learning is used to distinguish, for example, the sentences describing systemic stress from those describing cellular stress. A user account (free) is needed to create custom ontologies and save the search results. We do not track you.
Example: Rgma Penk.
By default, the addiction ontology is used. Custom ontologies can be created in user accounts.